FROGS Core

Galaxy
- Get Started ▾
- Reads processing
- Remove chimera
- Cluster/ASV filters
- Taxonomic affiliation
- Phylogenetic tree building
- ITSx
- Read demultiplexing
- Affiliatio filters
- Affiliation postprocessing
- Abundance normalisation
- Convert Biom file to TSV file
- Convert TSV file to Biom file
- Cluster/ASV report
- Affiliation report
Main tools
Optional tools

CLI
- Get Started
- Reads processing
- Remove chimera
- Cluster/ASV filters
- Taxonomic affiliation
- Phylogenetic tree building
- ITSx
- Read demultiplexing
- Affiliation filters
- Affiliation postprocessing
- Abundance normalisation
- Convert Biom file to TSV file
- Convert TSV file to Biom file
- Cluster/ASV report
- Affiliation report
Main tools
Optional tools
Affiliation statistics
Context
This is a bioinformatics tool used to generate metrics describing ASVs (Amplicon Sequence Variants) based on their taxonomic affiliations and the quality of these assignments.
It helps evaluate the reliability of taxonomic annotations, providing insights into the confidence of identifications, rarefaction curves, and taxonomic distribution within microbial communities.
How it does
The program reads a BIOM abundance file and metadata tags related to taxonomy and affiliation quality, such as bootstrap support, BLAST identity, and coverage.
It computes summary statistics for each taxonomic rank, evaluates rarefaction curves at selected ranks, and analyzes the confidence and consistency of taxonomic assignments.
This allows researchers to assess the quality of their taxonomic annotations and the diversity of their microbial samples.
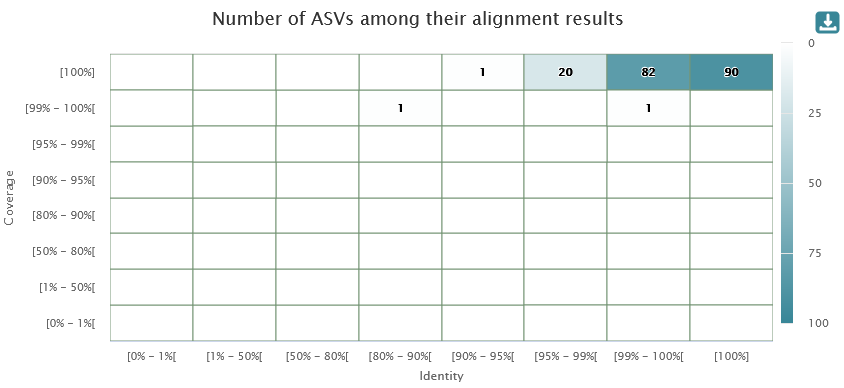
We obtain the same Taxonomy distribution and Alignment distribution HTML output as in the Taxonomic affiliation HTML output.
We obtain the same Taxonomy distribution and Alignment distribution HTML output as in the Taxonomic affiliation HTML output.
Configuration: 16S V3V4 Swarm
sbatch -J a_stats -o LOGS/affiliation_stats.out -e LOGS/affiliation_stats.err -c 8 --export=ALL --wrap="module load devel/Miniforge/Miniforge3 && module load bioinfo/FROGS/FROGS-v5.0.2 && affiliation_stats.py --input-biom FROGS/SWARM/affiliation.biom --log-file FROGS/affiliation_stats.log --html FROGS/SWARM/affiliation_stats.html --rarefaction-ranks Class Order Family Genus --multiple-tag 'blast_affiliations' --tax-consensus-tag 'blast_taxonomy' --identity-tag 'perc_identity' --coverage-tag 'perc_query_coverage' && module unload bioinfo/FROGS/FROGS-v5.0.2"
affiliation_stats.py --help)
Interpretation: 16S V3V4 Swarm
This report shows statistics of affiliation.
You can use the Krona output to explore the affiliation.
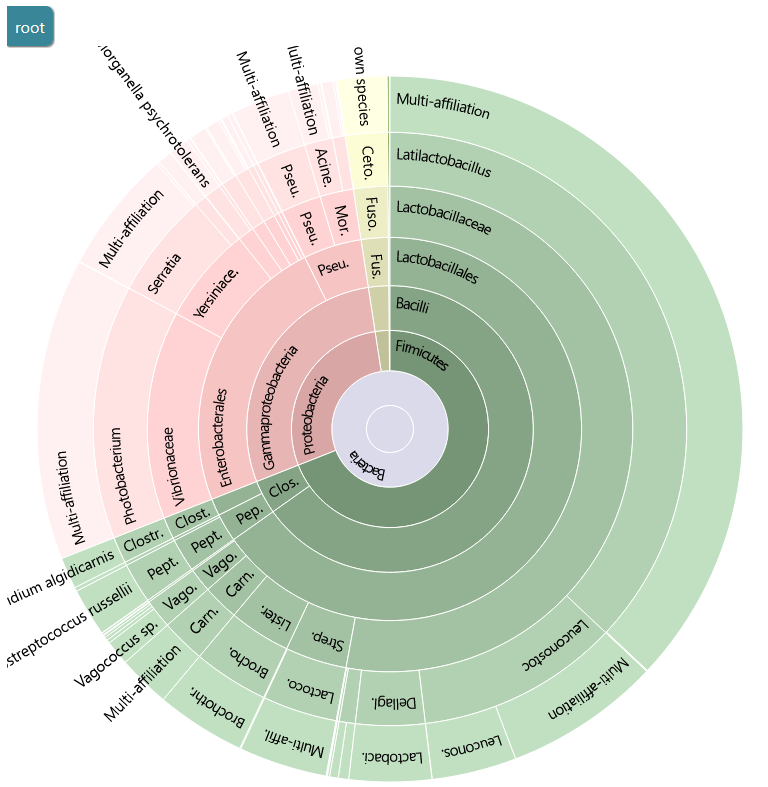
You can use the Krona output to explore the affiliation.


