FROGS Core

Galaxy
- Get Started ▾
- Reads processing
- Remove chimera
- Cluster/ASV filters
- Taxonomic affiliation
- Phylogenetic tree building
- ITSx
- Read demultiplexing
- Affiliatio filters
- Affiliation postprocessing
- Abundance normalisation
- Convert Biom file to TSV file
- Convert TSV file to Biom file
- Cluster/ASV report
- Affiliation report
Main tools
Optional tools

CLI
- Get Started
- Reads processing
- Remove chimera
- Cluster/ASV filters
- Taxonomic affiliation
- Phylogenetic tree building
- ITSx
- Read demultiplexing
- Affiliation filters
- Affiliation postprocessing
- Abundance normalisation
- Convert Biom file to TSV file
- Convert TSV file to Biom file
- Cluster/ASV report
- Affiliation report
Main tools
Optional tools
Affiliation Filters
Context
Once the clusters have been reconstructed and affiliated, it is sometimes useful to filter these data based on affiliation metrics or keywords.
This step is done in the FROGS Affiliation Filters tool
What it does
This tool removes or keeps ASVs or hides taxonomical metadata according to one or more criteria:
for RDP taxonomy : a minimal bootstrap threshold at a specific rank
for blast taxonomy : a minimal identity rate, coverage rate, or alignment length, or a maximal evalue, or the absence/presence of a full or partial taxon name.
Configuration: 16S V3V4 Swarm
We now apply filters to affiliation. For this example, we want to remove ASVs that are not Lactobacillales and ASVs that have a blast identity lower than 90.
sbatch -J filters -o LOGS/aff_filters.out -e LOGS/aff_filters.err -c 8 --export=ALL --wrap="module load devel/Miniforge/Miniforge3 && module load bioinfo/FROGS/FROGS-v5.0.2 && affiliation_filters.py --input-fasta FROGS/SWARM/filters.fasta --input-biom FROGS/SWARM/affiliation.biom --output-fasta FROGS/SWARM/affiliation_filters.fasta --log-file FROGS/SWARM/aff_filters.log --output-biom FROGS/SWARM/affiliation_filters.biom --html FROGS/SWARM/affiliation_filters.html --min-blast-identity 90 --delete --keep-blast-taxa Lactobacillales && module unload bioinfo/FROGS/FROGS-v5.0.2"
affiliation_filters.py --help)
Interpretation: 16S V3V4 Swarm
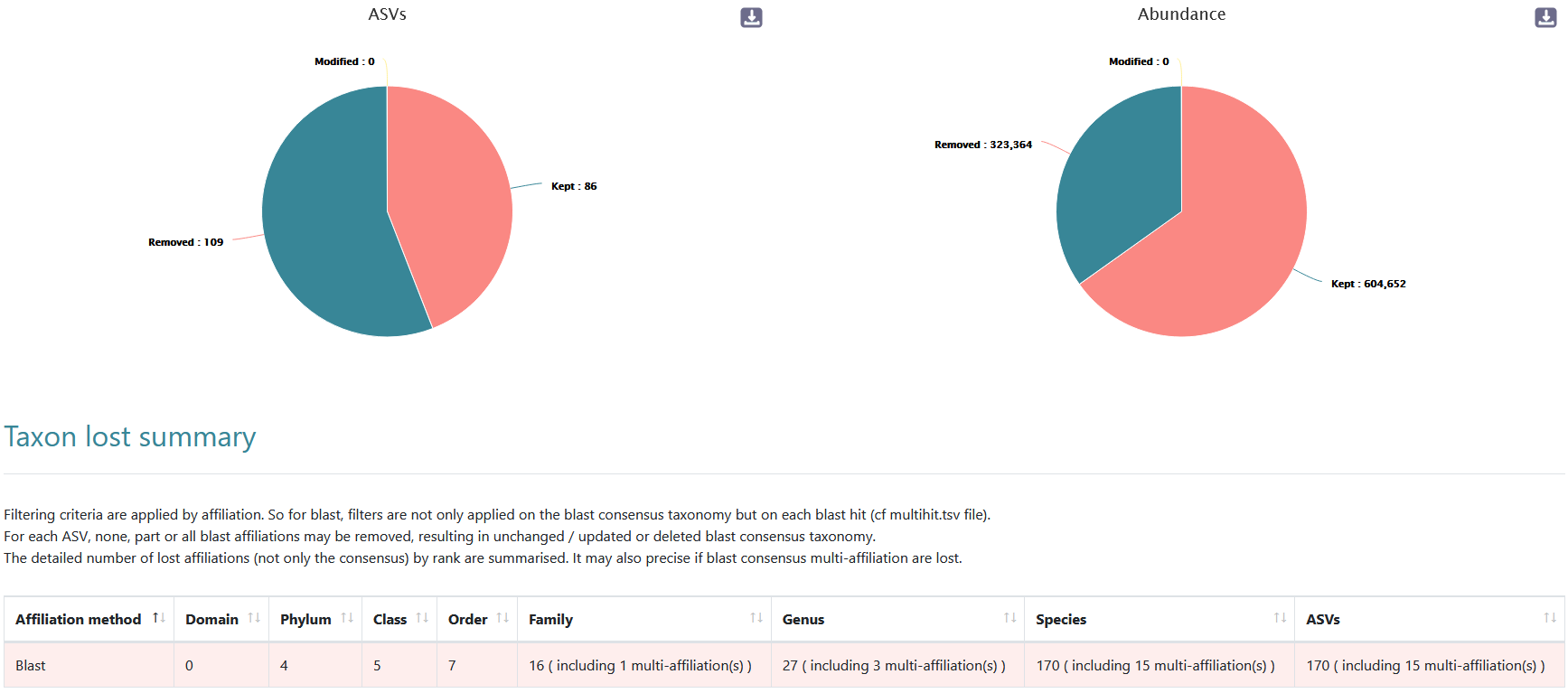
This report allows to show the impact of our filters:
- 109 ASVs are filtered out; 86 Lactobacillales are kept!
- ~35% of sequences are lost
- 604,652 sequences are remaining
- The Venn diagram shows that only one ASV was removed because it had less than 90% identity.
